1.1 E. coli as Model Organism
Studies using Escherichia coli as model organism have contributed most to our understanding of fundamental life processes (Piechocki 1989, Lee 2009, Samie 2017).
Figure 1: Escherichia coli as Model Organism
Escherichia coli, or short E. coli, is a Gram-negative, facultative anaerobic bacterium that commonly populates the intestine of homeothermic animals. Since its invention by the German-Austrian pediatrician Th. Escherich in 1885, E. coli has developed step by step into the molecular-biologically best studied and analyzed living system (Piechocki 1989, Idalia and Bernardo 2017). Apart from its cheapness, ease of cultivation and rapid population growth E. coli owes this success to the discovery of three processes by which individual bacteria of this species naturally exchange or can be forced to exchange genetic information (Piechocki 1989): (1) conjugation, (2) transduction and (3) transformation.
Ad (1): Conjugation refers to the juxtaposition of two bacteria to exchange extra-chromosomal DNA localized in an F-Plasmid via a fusing organism bridge. This represents a para-sexual phenomenon among bacteria. The process was discovered on E. coli by J. Lederberg and E. Tatum as early as 1946.
Ad (2): Transduction is the injection and transfer of DNA among bacteria via bacteriophages discovered on E. coli by Zinder and the Lederbergs several years later (Lederberg 1950, Zinder and Lederberg 1952).
Ad (3): Transformation is a further variant of gene transfer in which merely DNA fragments are taken up by a bacterium. This variant is known since 1944.
This genetic analysis tool set enabled scientists to address and solve a plethora of fundamental questions of life as listed here (Piechocki 1989): How are genes replicated? How are genes structured? How is the genetic information encoded? How is the genetic information expressed? How do mutations occur? How is the activity of genes regulated? In consequence, scientific work conducted on E. coli has been rewarded with 12 Nobel Prizes.
Table 1: Nobel Prizes based on extensive usage of Escherichia coli (modified from https://microbeonline.com, descriptions from https://www.nobelprize.org/)
Year | Nobel Laureate(s) | Nobel Prize on/in | for |
1958 | Joshua Lederberg,
George Wells Beadle & Edward Tatum | Physiology or Medicine | his discoveries concerning genetic recombination and the organization of the genetic material of bacteria their discovery that genes act by regulating definite chemical events |
1959 | Severo Ochoa & Arthur Kornberg | Physiology or Medicine | their discovery of the mechanisms in the biological synthesis of ribonucleic acid and deoxyribonucleic acid |
1965 | François Jacob, André Lwoff & Jacques Monod | Physiology or Medicine | their discoveries concerning genetic control of enzyme and virus synthesis |
1968 | Robert W. Holley, Har Gobind Khorana & Marshall W. Nirenberg | Physiology or Medicine | their discoveries concerning the molecular structure of nucleic acids and its significance for information transfer in living material |
1969 | Max Delbrück, Alfred D. Hershey & Salvador E. Luria | Physiology or Medicine | their discoveries concerning the replication mechanism and the genetic structure of viruses |
1978 | Werner Arber, Daniel Nathans & Hamilton O. Smith | Physiology or Medicine | the discovery of restriction enzymes and their application to problems of molecular genetics |
1980 | Paul Berg | Chemistry | his fundamental studies of the biochemistry of nucleic acids, with particular regard to recombinant-DNA |
1989 | Sidney Altman & Thomas R. Cech | Chemistry | their discovery of catalytic properties of RNA |
1997 | Paul D. Boyer & John E. Walker | Chemistry | their elucidation of the enzymatic mechanism underlying the synthesis of adenosine triphosphate (ATP) |
1999 | Ahmed H. Zewail | Chemistry | his studies of the transition states of chemical reactions using femtosecond spectroscopy |
2008 | Osamu Shimomura, Martin Chalfie & Roger Y. Tsien | Chemistry | the discovery and development of the green fluorescent protein, GFP |
2015 | Tomas Lindahl, Paul Modrich & Aziz Sancar | Chemistry | mechanistic studies of DNA repair |
2020 | Emmanuelle Charpentier & Jennifer A. Doudna | Chemistry | the development of a method for genome editing |
In particular, the discovery of restriction enzymes by W. Arber, D. Nathans & H. O. Smith (Nobel prize 1978) suddenly opened up a new experimental strategy for both, basic and practical research, in which DNA fragments from any organism, even from higher organisms, could be fused together and transferred into the bacterial cell by way of suitable vectors (plasmids or phages, see the method set above). Thus, Escherichia coli became the “factory” for the propagation of any gene. Thereby, microbial genetics, molecular biology and genetic engineering opened the way to bio-technological applications and ushered in a new industrial revolution (Piechocki 1989).
For instance, two of the world’s largest insulin-producing firms (Eli Lilly and Company, Sanofi) that have together a market share of about 54% (insulin-volume based, 2016, from https://marketrealist.com), rely on E. coli as host for their manufacturing of human insulin. In 2019 463 million people worldwide were living with diabetes (https://www.diabetesatlas.org). 5 – 10%, hence 23.15 – 46.3 million have type 1 (insulin-dependent) diabetes and 18.7% of the remaining 416.7 – 439.9 million, thus 77.9 – 82.3 million people, are type 2 diabetics who are treated with insulin (Basu et al. 2019, 18.7% is the mean % of type 2 diabetics treated with insulin across all studies mentioned therein, weighted by the population of the county in which each study took place).
Thus, we can assess that 54.6 – 69.4 million people worldwide rely on obtaining insulin produced by Escherichia coli to survive.

Figure 2: A fermenter for the cultivation of genetically engineered bacteria (National Museum of American History, from https://www.smithsonianmag.com)
Because of the outstanding role of Escherichia coli as model organism, sequencing of its whole genome was suggested as early as 1983 (Blattner 1983), but it took until 1997 that the genome project was finalized. The first to be completed was the sequencing of the strain K12 (Blattner et al. 1997), the most widely used strain in laboratories around the world, and since then many strains have followed. This has certainly helped to stimulate further research to gain a more thorough understanding of the bacterium. E. coli is now serving as a platform system for evolutionary ecology (Lenski 2017, Blount et al. 2018) and systems biology studies (Lee 2009).
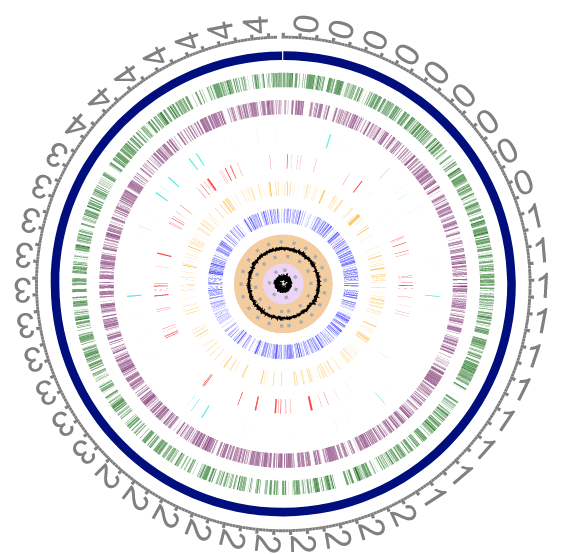
Figure 3: The circular genome of Escherichia coli str. K-12 substr. MG1655 (from https://patricbrc.org)
The E. coli Long-Term Evolution Experiment (LTEE) is probably the most well-known study of the former category. Since its inception in 1988, the researchers around Richard Lenski have monitored and analyzed dynamics of the adaption by natural selection in 12 initially identical populations of an ancestral asexual E. coli strain in a glucose-limited minimal salts medium with daily 100-fold dilution of the grown populations. In 2020 the populations had reached 73,500 generations. Several unexpected discoveries were being made during the experiment (Lenski 2017, Blount et al. 2018): After 50,000 generations the populations grew 70% faster than their ancestors. The relative fitness improvement slowed down over time but fitness is prognosed to rise forever (following a power-law trajectory). Some populations diverged into two stably coexisting ecotypes, one that uses glucose and produces acetate as metabolic by-product (but has lost the ability to use acetate itself), whereas the other uses acetate as carbon source. Hypermutability with increased probability of beneficial mutations and higher risk of deleterious mutations alike developed severalfold. One population even gained the ability to aerobically consume citrate as carbon source.

Figure 4: The 12 populations propagated in the E. coli LTEE
Systems metabolic engineering is a rapidly advancing application area that belongs to the latter category (Chaper 20 in Lee 2009). It is powered by systems-biological tools and methods. Systems metabolic engineering aims to develop superior strains suitable for the industrial-level production of bioproducts. First, genome engineering is used to construct a base strain with improved biosynthetic performance from the chosen wild-type strain. Common methods applied during this step are: amplification of biosynthetic genes, removal of negative regulations, deletion of competing pathways and whole genome minimization. For the construction of the ultimate superior strain a combination of “omics” analysis (including genomics, transcriptomics, proteomics, metabolomics, fluxomics) and computational analysis of metabolites and fluxes in metabolic networks is applied. Finally, factors of the bio-technological fermentation process, such as removal of byproducts, medium optimization and purification are considered in systems metabolic engineering. A striking recent application of systems metabolic engineering developed by Sang Yup Lee’s group is the fermentative production of aromatic polyesters from glucose by engineered E. coli strains as potential substitute for petroleum-based plastic production in the future (Yang et al. 2018).

Figure 5: Cellular Metabolism Overview Escherichia coli K-12 substr. MG 1655 (from https://www.ecocyc.org)
Another research area on which systems biology has focused for long is the behavioral response of E. coli bacteria to environmental signals on the whole-cell level (chemotaxis) and the multicellular level (biofilm formation) via the sensory transduction network (Chapter 8 in Lee 2009). Simulation models have played a crucial role in the experimental elucidation of the signal transduction mechanisms. We will elaborate on both aspects in the following sections.
Literature Cited
Basu, S., J. S. Yudkin, S. Kehlenbrink, J. I. Davies, S. H. Wild, K. J. Lipska, and J. B. Sussman, et al. 2019. Estimation of global insulin use for type 2 diabetes, 2018–30: a microsimulation analysis. The Lancet Diabetes & Endocrinology. 7:25–33.
Blattner, F. R. 1983. Biological frontiers. Science. 222:719–720.
Blattner, F. R., G. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, and J. Collado-Vides, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science. 277:1453–1462.
Blount, Z. D., R. E. Lenski, and J. B. Losos. 2018. Contingency and determinism in evolution: Replaying life's tape. Science (New York, N.Y.). 362.
Idalia, V.-M. N., and F. Bernardo. 2017. Escherichia coli as a Model Organism and Its Application in Biotechnology in A. Samie, ed. Escherichia coli - Recent Advances on Physiology, Pathogenesis and Biotechnological Applications. InTech.
Lederberg, E. Z. 1950. Lysogenicity in Escherichia coli strain K-12. Microbial Genetics Bulletin. 1:6–8.
Lee, S. Y. 2009. Systems biology and biotechnology of Escherichia coli. Springer, Dordrecht.
Lenski, R. E. 2017. Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. The ISME journal. 11:2181–2194.
Piechocki, R. 1989. Das berühmteste Bakterium. 100 Jahre Escherichia-coli-Forschung. Aulis Verl., Köln.
Samie, A., ed. 2017. Escherichia coli - Recent Advances on Physiology, Pathogenesis and Biotechnological Applications. InTech.
Yang, J. E., S. J. Park, W. J. Kim, H. J. Kim, B. J. Kim, H. Lee, and J. Shin, et al. 2018. One-step fermentative production of aromatic polyesters from glucose by metabolically engineered Escherichia coli strains. Nature Communications. 9:79.
Zinder, N. D., and J. Lederberg. 1952. Genetic exchange in Salmonella. Journal of bacteriology. 64:679–699.
